Abstract Text
We introduce a Bayesian genetic algorithm for reconstructing atomic models of catalytic nanoparticles from a single projection using Z-contrast imaging. Atom-counting results obtained from annular dark field scanning transmission electron microscopy (ADF STEM) images serve as an input for the initial three-dimensional (3D) model. The novel algorithm minimizes the energy of the structure while exploiting a priori information about the imprecision of the atom-counting results. The results show good prospects for obtaining reliable reconstructions from images acquired with lower incident electron doses.
In order to fully exploit structure-property relations of nanomaterials, a 3D characterization at high resolution is often required. In recent years, the resolution of electron tomography has reached the atomic scale. However, many interesting materials are too beam-sensitive to handle the substantial electron dose of the multiple projections required for electron tomography. Therefore, an alternative method has been developed where the 3D atomic structure is reconstructed from atom counts obtained from a single ADF STEM projection image. These atom counts can be used to create an initial atomic model which serves as an input for an energy minimization to obtain a relaxed 3D reconstruction of a nanostructure [1-3].
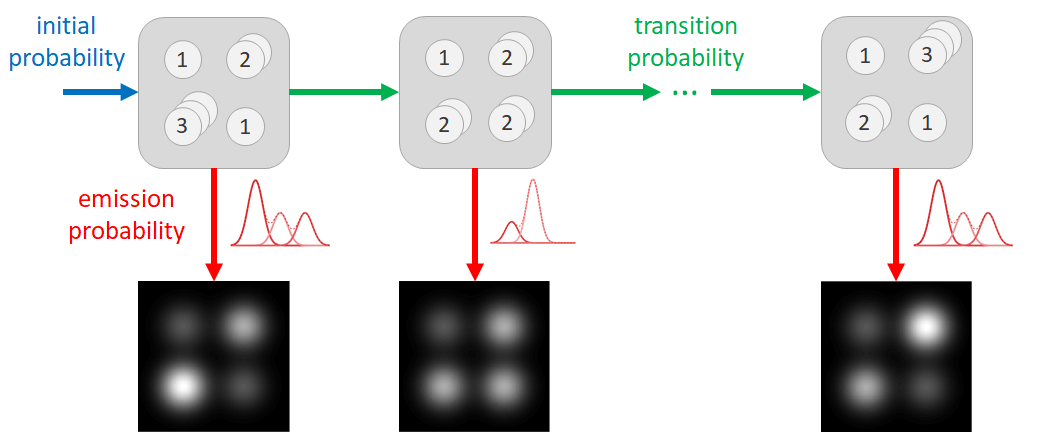
Two possible approaches for the energy minimization are nowadays available. Using the first approach, the energy is minimized by shifting the atomic columns up and down while keeping the number of atoms in a column fixed to the outcome of the atom-counting procedure [2]. The second approach consists of a full molecular dynamics simulation to relax the particle’s structure [3]. Where the first method is too strict by ignoring atom-counting imprecision, especially at lower doses, the second method runs the risk of ending up in a global energy minimum and violating the physical constraints of the experimental observation. To overcome these limitations, we propose to include the atom-counting imprecision via a Bayesian inference scheme. Moreover, the incorporation of additional prior knowledge from neighbour-mass relations will be beneficial when reconstructing atomic models from low dose ADF STEM images. Genetic algorithms (GA) are powerful tools for solving such large optimization problems [4,5]. Here, our prior knowledge is fused into a new Bayesian GA for reconstructing the 3D atomic structure.
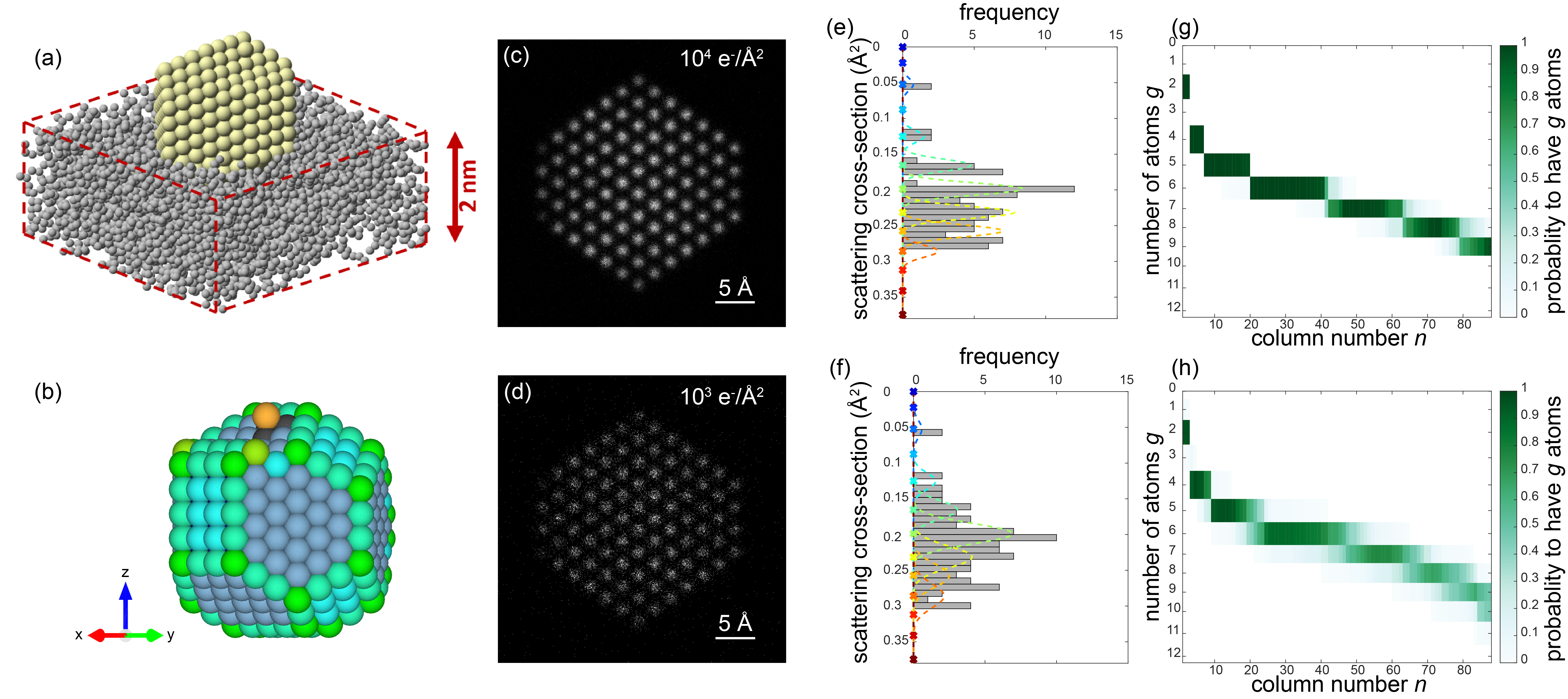
Figures 1(a-b) show the model of the Pt nanoparticle embedded in a carbon support which is used in our simulation study. Figures 1(c-d) illustrate simulated ADF STEM images for 2 different electron doses. By modelling the images as a superposition of Gaussian peaks located at the atomic columns, the volume under each peak can be estimated by fitting this model to the region of interest. These volumes are integrated intensities of electrons and thus correspond to scattering cross-sections. In a subsequent analysis, the distribution of the scattering cross-sections of all atomic columns is decomposed into overlapping normal distributions as illustrated in Figures 1(e-f) [6]. The number of atoms in each projected atomic column is then obtained by assigning the scattering cross-section to the component that generates this scattering cross-section with the highest probability. The width of the distributions in Figures 1(e-f) reflects the imprecision on the atom-counting results and can be used to define the probability for an atomic column to contain a specific number of atoms (Figures 1(g-h)). This probability is included as a priori information in the energy minimization scheme for the 3D atomic reconstruction of the nanoparticle. In this manner, the removal or addition of an atom to a column is only allowed within the finite precision of the atom-counting result. In addition to the a priori knowledge from the atom-counting imprecision, we include also neighbour-mass relations as further a priori knowledge that abrupt discontinuities are highly non-physical.
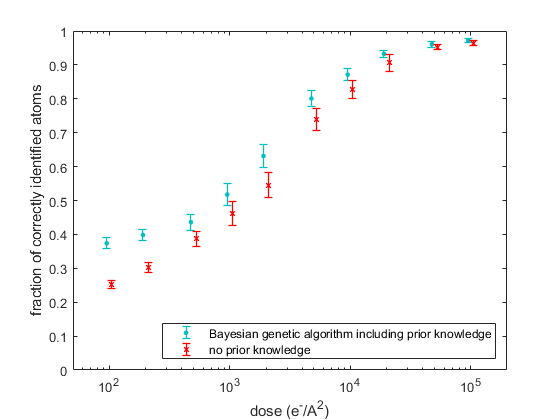
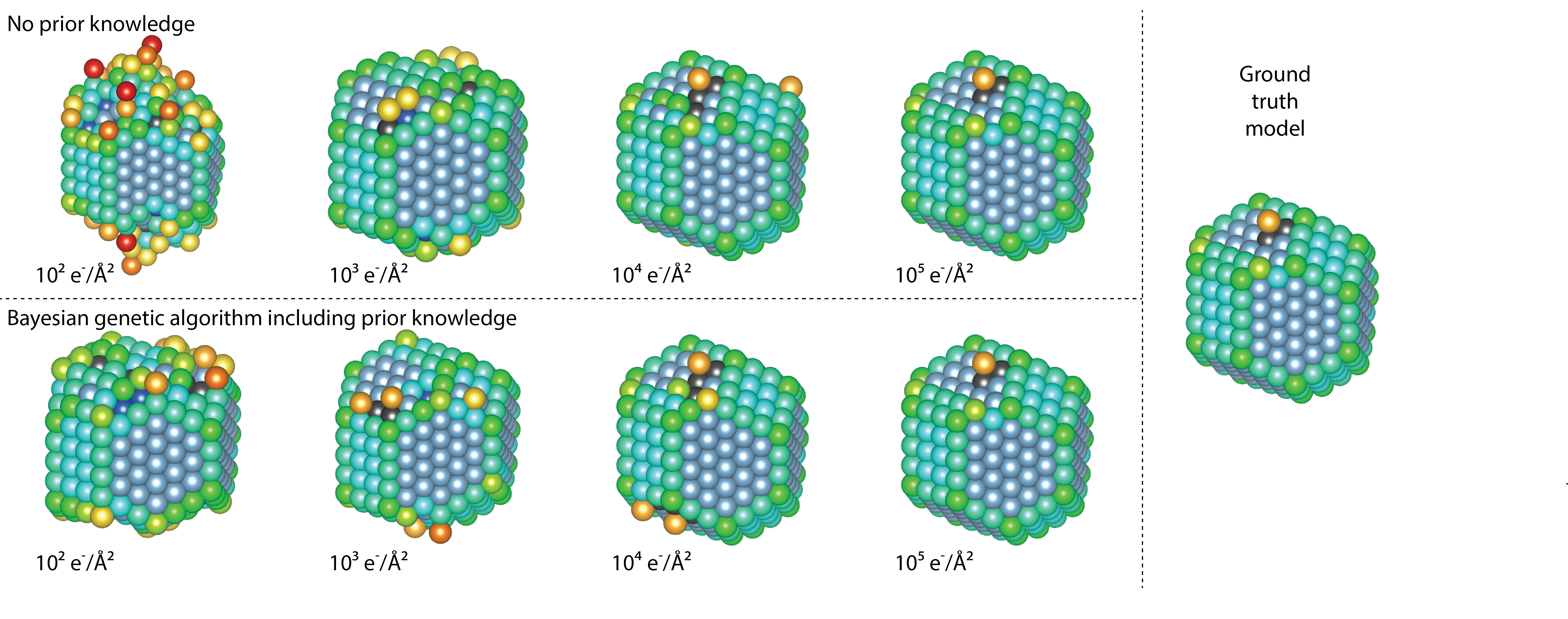
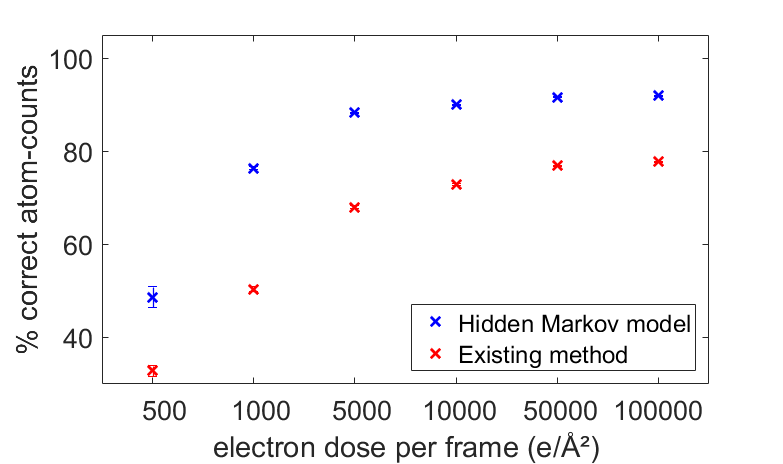
In an extensive simulation study, the quality of the obtained reconstructions is quantitatively evaluated in terms of the coordination number of the surface atoms, which are of general interest for catalysis. Figure 2 shows the fraction of the surface atoms with the same coordination number as the ground truth Pt model. As a reference, the results for the reconstructions without prior knowledge are also included. Reconstructed models at different doses and the ground truth model are displayed in Figure 3. From Figures 2 and 3, it is clear that a significant improvement is observed when including more relevant prior knowledge, especially at lower doses.
In conclusion, an advanced Bayesian genetic algorithm to reconstruct 3D atomic models of nanoparticles will be presented with its possibilities and limitations. The method is very promising for obtaining reliable 3D atomic models of beam-sensitive nanoparticles during dynamical processes from images acquired with lower incident electron doses [7].
Figure 1 (a) Model for a Pt nanoparticle embedded in the carbon support. (b) Model of the same Pt nanoparticle where the coloring of the Pt atoms indicates the nearest-neighbor coordination. (c-d) Simulated ADF STEM images of the nanoparticle in (a) along the 110 axis for incident electron doses of 104 e-/Å2 and 103 e-/Å2. (e-f) Distribution of the scattering cross-sections for the two incident electron doses of 104 e-/Å2 and 103 e-/Å2 (g-h) corresponding probabilities for a column n to contain a specific number of atoms g.
Figure 2 Fraction of the correctly identified surface atoms with the same coordination number as the ground truth Pt model as a function of the incident electron dose.
Figure 3 Ground truth model and reconstructed models without and with prior knowledge at different doses. The coloring of the Pt atoms indicates the nearest-neighbour coordination from 1 in red to 12 in dark blue.
Keywords
3D atomic modelling
atom-counting
Bayesian inference
genetic algorithm
References
[1] S. Bals et al., Nature Communications 3 (2012), p. 897.
[2] L. Jones et al., Nano Letters 14 (2014), p. 6336.
[3] A. De Backer et al., Nanoscale 9 (2017), p. 8791.
[4] M. Yu et al., ACS Nano 10 (2016), p. 4031.
[5] Aarons et al., Nano Letters 17 (2017), p. 4003.
[6] A. De Backer et al., Ultramicroscopy 134 (2013), p. 23.
[7] This work was supported by the European Research Council (Grant 770887 PICOMETRICS to SVA and Grant 823717 ESTEEM3). The authors acknowledge financial support from the Research Foundation Flanders (FWO, Belgium) through project fundings and a postdoctoral grant to ADB. LJ acknowledges Science Foundation Ireland (SFI), the Royal Society, and the AMBER Centre.