Abstract
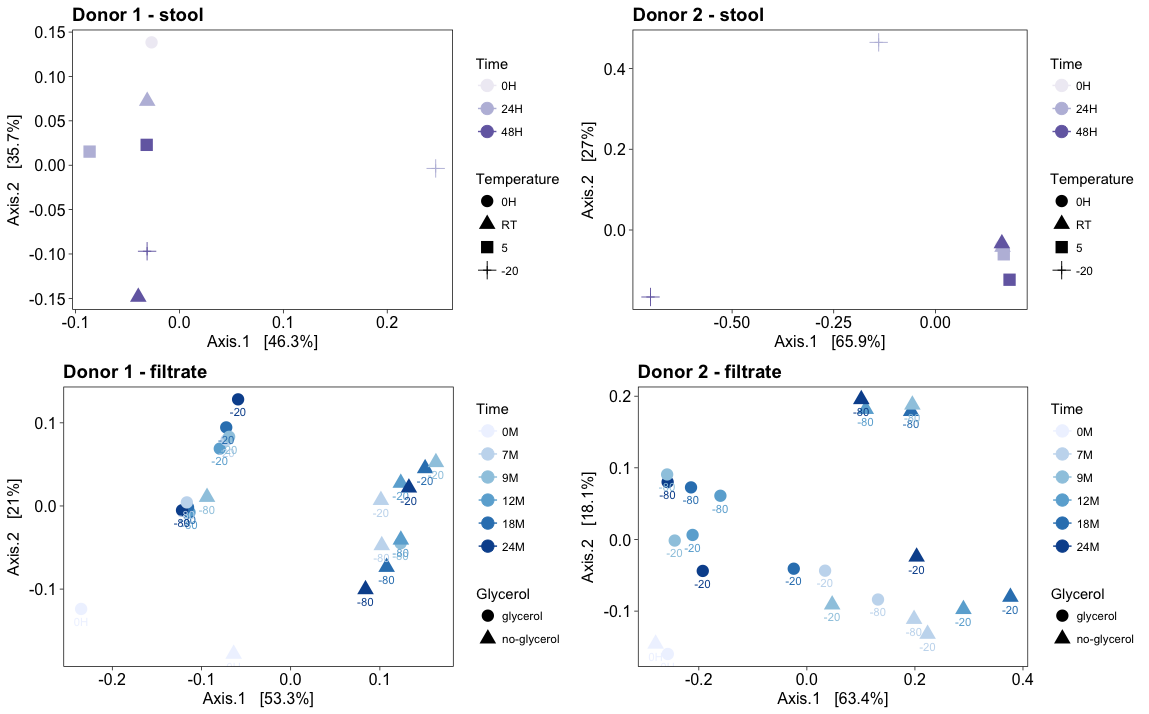
Objectives: Stool storage is key to researching the association between gastrointestinal dysbiosis and disease states. Fecal microbiota transplantation using frozen filtrate is used for patients with recurrent Clostridioides difficile infection. This study determined the impact of storing stool and frozen filtrate on microbiome composition. Methods: Fresh stool was obtained from a high-diversity (HD) and low-diversity (LD) donor. Aliquots were stored at room temperature (RT), 5C, and at -20C for 24 and 48 hours, or processed immediately. Fresh stool was also homogenized with both 0.9N-sterile-saline and 0.9N-sterile-saline containing 10%-glycerol. Resulting filtrate aliquots were frozen at -20C and at -80C. At baseline and after 7, 9, 12, 18 and 24M storage, gDNA was isolated using the MO BIO PowerSoil® DNA Isolation Kit. 16s rRNA gene amplicon sequencing targeting the V4 hypervariable region was performed on the Illumina MiSeq. Low abundant OTUs were excluded. Results: Differences in microbiota profiles were observed in both HD and LD stool when stored immediately at –20C, more so with the LD stool. Differences were also noted for LD stool stored at room temperature and less so at 5°C. Storage at RT or 5°C had no impact for the HD stool. Long-term filtrate storage of the HD stool filtrate at –80C with 10%-glycerol best preserved the bacterial microbiota profile followed by –20C with 10%-glycerol then –80C and –20C without 10%-glycerol. Differences were also observed in the LD stool filtrate when stored in any conditions, with the least impact being –80°C with glycerol. Conclusions: To preserve microbiota profiles, storing stool at 5°C until received in the laboratory is optimal. For researchers assessing microbiota correlations with disease states, immediate gDNA extraction from stool upon receipt is ideal. For those using frozen filtrate from healthy donors for fecal transplants, long-term storage is best at –80C with 10%-glycerol.